Standardize your cfDNA-based NGS assays with Seraseq® ctDNA Controls
The most patient-like reference materials for cfDNA NGS assays
Expedite the clinical implementation and standardization of next-generation sequencing (NGS)-based liquid biopsy assays with patient-like ctDNA reference samples featuring broad coverage of clinically relevant genomic alterations. Trust the industry-leading portfolio of Seraseq® circulating tumor DNA (ctDNA) reference materials to support the analytical validation and clinical deployment of your cell free DNA NGS-based assays.
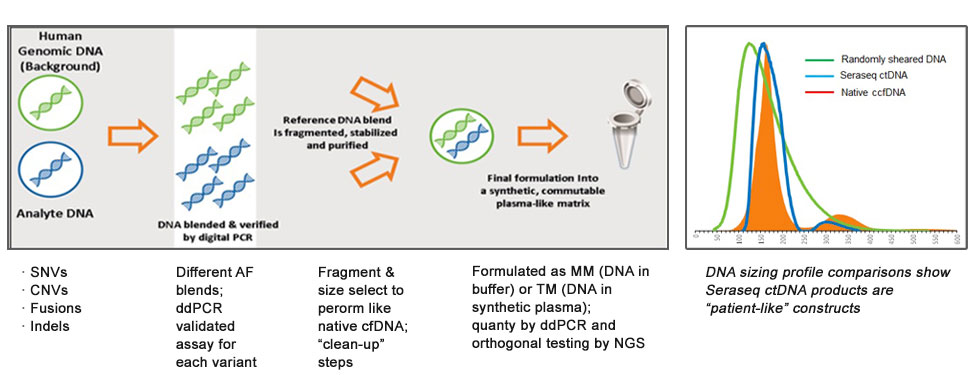
Figure-1: A generic workflow for the development of Seraseq ctDNA reference materials. Insert picture highlights the patient-like profile of the Seraseq ctDNA fragment sizes when compared to that of natural circulating cell free DNA (native ccfDNA).
Our Patent-Pending1 ctDNA Technology Enables
- Easy access to the broadest range of clinically relevant DNA biomarkers
- Comprehensive analytical validations and LoD determinations with range of AFs
- Natural cfDNA-like size distribution and performance
- Offered in purified (Mutation Mix) and encapsulated (Plasma-mimetic) formats
- Available as pre-mixed content or customize to suit your assay or clinical needs
- Use as QC for plasma sample extraction efficiency and end-to-end assay workflow optimization
Comprehensive Solutions Across All Phases of The NGS Workflow
Seraseq® ctDNA v2 controls
-
- Pan-cancer (solid tumor & heme disorder) profiling/analysis
- Comprehensive ctDNA reference sample content
- 28 genes and 40 variants - SNVs, INDELs, and SVs
- AF levels from 0.125% to 2%, including WT
- Purified mutation mix (MM) and synthetic plasma (RM) formats
Gene Table
| AKT1● | CTNNB1● | FLT3● | GNAS● | GNAS● | NRAS/CSDE1● | RET● |
| APC●◊ | EGFR●◊ | FOXL2● | IDH1● | MPL● | PDGFRA●◊ | SMAD4◊ |
| ATM◊ | ERBB2◊ | GNA11● | JAK2● | NCOA4-RET ‡ | PIK3CA●◊ | TP53●◊ |
| BRAF● | FGFR3● | GNAQ● | KIT● | NPM1◊ | PTEN●◊ | TPR-ALK‡ |
● = SNV ◊ = INDEL ‡ = SV
Product Listing
| Format | Material # | AF | Total Mass | Link | Format | Material # | AF | Total Mass | Link |
Synthetic Plasma Matrix |
0170-0203 | 2% | 125 ng | Click Here | Purified Mix |
0710-0139 | 2% | 250 ng | Click Here |
| 0710-0204 | 1% | 125 ng | Click Here | 0710-0140 | 1% | 250 ng | Click Here | ||
| 0710-0205 | 0.5% | 125 ng | Click Here | 0710-0141 | 0.5% | 250 ng | Click Here | ||
| 0710-0207 | 0.125% | 125 ng | Click Here | 0710-0142 | 0.25% | 250 ng | Click Here | ||
| 0710-0208 | WT | 125 ng | Click Here | 0710-0143 | 0.125% | 250 ng | Click Here | ||
| 0710-0144 | WT | 250 ng | Click Here |
Seraseq® ctDNA Complete™ controls
- Content focused on solid tumor disease variants
- Coverage of all clinically-actionable / targeted variants in a single reference sample
- 16 genes and 25 variants - SNVs, INDELs, CNVs, and SVs
- AF levels from 0.1% to 5%, including WT
- Offered as both Mutation Mix (MM) and synthetic plasma (RM) formats
Gene Table
| AKT1● | BRCA2◊ | ERBB2▪◊ | MYC▪ |
| ALK ● | CD74-ROS1‡ | KIT● | NCOA4-RET‡ |
| BRAF ● | EGFR●◊ | KRAS● | NRAS● |
| BRCA1◊ | EML4-ALK‡ | MET ▪ | PIK3CA●◊ |
● = SNV ◊ = INDEL ▪ = CNV ‡ = SV
Product Listing
| Format | Material # | AF | Total Mass | Link | Format | Material # | AF | Total Mass | Link |
Synthetic Plasma Matrix |
0170-0669 | 5% | 125 ng | Click Here | Purified Mix |
0710-0528 | 5% | 250 ng | Click Here |
| 0710-0670 | 2.5% | 125 ng | Click Here | 0710-0529 | 2.5% | 250 ng | Click Here | ||
| 0710-0671 | 1% | 125 ng | Click Here | 0710-0530 | 1% | 250 ng | Click Here | ||
| 0710-0672 | 0.5% | 125 ng | Click Here | 0710-0531 | 0.5% | 250 ng | Click Here | ||
| 0710-0673 | 0.1% | 125 ng | Click Here | 0710-0532 | 0.1% | 250 ng | Click Here | ||
| 0710-0674 | WT | 125 ng | Click Here | 0710-0533 | WT | 250 ng | Click Here |
Seraseq ctDNA Quantitation
Seraseq ctDNA reference materials are derived from synthetic plasmids where all variants are blended at idealized allelic frequencies. These reference materials are then quantitated using digital droplet PCR (ddPCR), and orthogonally analyzed by a cfDNA-based targeted NGS assay (see, e.g., Figure 2).
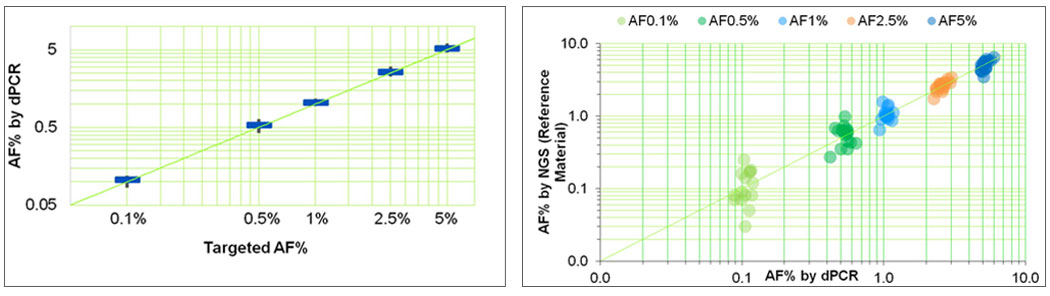
Figure-2: Analysis of the 25 variants in the Seraseq ctDNA Complete™ reference material at blended AF levels (0.1%, 0.5%, 1%, 2.5% and 5%) by (a) digital PCR (ddPCR) and (b) concordance between targeted cfDNA NGS assay (Archer Dx’s Reveal-28 ctDNA) and ddPCR (Bio-Rad Lab’s QX200)2
Multi-Lab Evaluation of Seraseq ctDNA Reference Materials
Under the auspices of the National Institute of Standards and Technology (NIST), a multi-laboratory study was conducted to ascertain the utility and viability of contrived reference materials such as the Seraseq® ctDNA v2 reference material (plasma and mutation mi formats) as an effective surrogate cfDNA sample in cancer patient analysis. Laboratories involved in the evaluation include NIST, John’s Hopkins University, Frederick National Laboratory for Cancer Research, Boston University, University of North Carolina, University of Gotenburg’s Sahlgrenka Cancer Center and Wallenberg Center for Molecular and Translational Medicine, the Early Detection Research Network at the NCI, as well as commercial diagnostic labs at Asuragen, Inc., Archer Dx, and SeraCare Life Sciences.
The study evaluated the Seraseq ctDNA v2 reference material in plasma extraction efficiencies, cfDNA fragment sizing, blended AF accuracy, as well as precision and sensitivity of variant detection in molecular cfDNA assays using digital droplet PCR and targeted NGS chemistries (see, e.g., Figure 3). Results of the study were published in the Journal of Molecular Diagnostics (2019).3
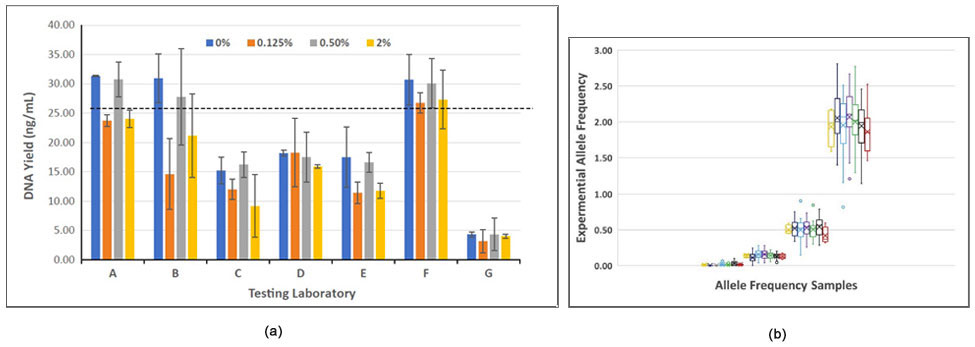
Figure-3: (a) DNA yields from different extraction chemistries versus expected (dotted line), and (b) Comparison of NGS results of analysis of blended AF versus observed AF values by all 7 laboratories.
Introducing Two New Industry-firsts Liquid Biopsy Reference Controls—bTMB and MRD Products
Seraseq® Blood TMB Mix - Blood Tumor Mutational Burden
Blood TMB (bTMB) combines all of the challenges of ctDNA assays with all of the challenges of assessing TMB with targeted gene panels. Only a handful of diagnostic laboratories presently measure bTMB, and their analytic validations rely on hard-to-obtain specimens. This specimen scarcity and frequent lack of a “true” TMB score impedes in-depth analytic assay validation and the generation of harmonized bTMB data. There are also reports of discordance in bTMB scores and whether bTMB is a relevant biomarker for checkpoint inhibitors.
Assessment of bTMB Scores in 2 Targeted ctDNA NGS Assays
For bTMB measurements to become routine, robust, and accessible to all diagnostic laboratories, we developed contrived patient-like bTMB reference materials.7 These samples were assessed by two independent targeted ctDNA NGS assays at TMB Score 7, 9, 20, 26 for the corresponding bTMB Scores. Results are shown in Figure 4 below.8
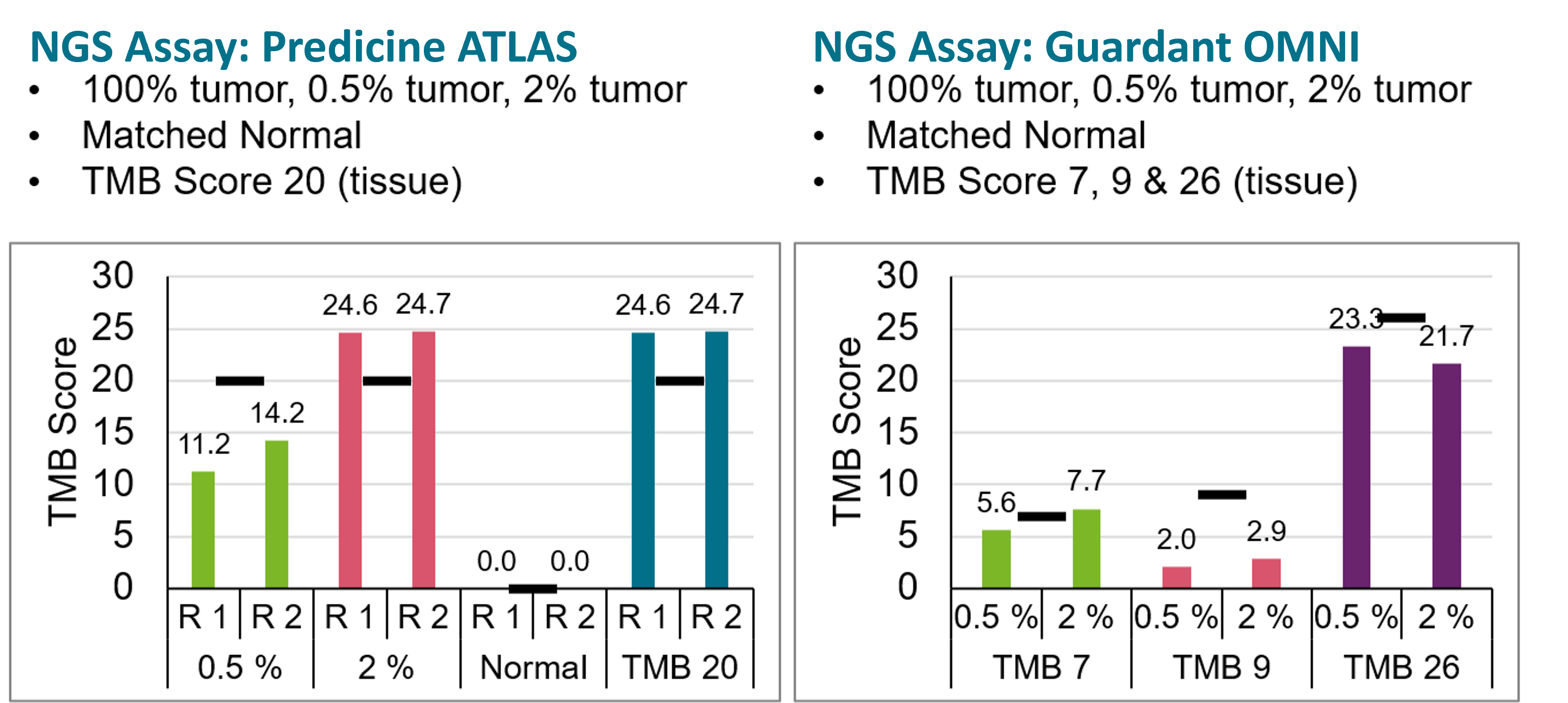
Figure-4: Measured bTMB Scores at 0.5% and 2% tumor content in two ctDNA NGS assays, and comparison to WES-based () tissue TMB Scores.8
Seraseq® MRD Mix for Minimal Residual Disease Monitoring
The analytical validation of liquid biopsy-based NGS assays for monitoring the disappearance and reemergence of cancer can be challenging without use of reference materials that allow for the assessment of sensitivity and specificity at variant allele frequencies (VAFs) over an order of magnitude below those that can be detected reliably by typical circulating tumor DNA (ctDNA) NGS assays. At such low VAFs, a given plasma sample may only contain limited somatic variants - and only at a few copies - which is why some MRD assays are now focusing on monitoring patient-specific somatic variants throughout the genome using patient-specific custom assays. This also means that validation should include the steps needed to monitor patient-specific somatic variants.
The new Seraseq ctDNA MRD reference material was designed for a tumor/normal workflow and consists of three components:9
(1) A normal DNA cell line to assess specificity.
(2) A germline SNP-matched cell line with hundreds to thousands of additional somatic variants.
(3) Blended, fragmented and sized DNA to mimic circulating cell-free DNA (ccfDNA) and serve as the input for MRD assays at VAFs from 1% to 0.001% (one mutant copy per 0.35 to 350 ng of ccfDNA) and at 0%.
For more details about this product or any Seraseq product, please contact us by phone +1 800-676-1881 or by email to infobox@seracare.com.
For more information on liquid biopsy reference materials or to have someone contact you, please fill out the form below.
References
1. Konigshofer, Y. Methods for Preparing DNA Reference Material and Controls. United States; WO 2018/094183 Al, 2018
2. Ruminski-Lowe, D., et. al., New technology to generate commutable and comprehensive circulating tumor DNA (ctDNA) reference materials for next generation sequencing, Poster#: TT034. 2018 AMP Meeting, San Antonio, TX, USA.
3. Hua-Jun He, Erica V. Stein, Yves Konigshofer, Thomas Forbes, Farol L. Tomson, Russell Garlick, Emiko Yamada, Tony Godfrey, Toshiya Abe, Koji Tamura, Michael Borges, Michael Goggins, Sandra Elmore, Margaret L. Gulley, Jessica L. Larson, Lando Ringel, Brian C. Haynes, Chris Karvolich, P. Mickey Williams, Aaron Garnett, Anders Stahlberg, Stefan Filges, Lynn Sorbara, Mathew R. Young, Sudhir Srivastava, and Kenneth D. Cole. MultiLaboratory Assessment of a New Reference Material for Quality Assurance of Cell Free Tumor DNA Measurements. J. Mol. Diag., 2019, Vol 21, No 4, 658-676 (https://www.jmdjournal.org/article/S1525-1578(18)30479-3/fulltext).
4. Sabrina Weber, Benjamin Spiegl, Samantha O. Perakis, Christine M. Ulz, Peter M. Abuja, Karl Kashofer, Paul van der Leest, Maria Aguirre Azpurua, Menno Tamminga, Dan Brudzewsky, Dominic G. Rothwell, Sumitra Mohan, Alexander Sartori, Rita Lampignano, Yves Konigshofer, Markus Sprenger-Haussels, Harriet Wikman, Inger R. Bergheim, Vera Kloten, Ed Schuuring, Michael R. Speicher and Ellen Heitzer Technical Evaluations of Commercial Mutation Analysis Platforms and Reference Materials for Liquid Biopsy Profiling, Cancers, 2020, 12, 1588 (doi:10.3390/cancers12061588)
5. Brandon R. McDonald, Tania Contente-Cuomo, Stephen-John Sammut, Ahuva Odenheimer-Bergman, Brenda Ernst, Nieves Perdigones, Suet-Feung Chin, Maria Farooq, Rosa Mejia, Patricia A. Cronin, Karen S. Anderson, Heidi E. Kosiorek, Donald W. Northfelt, Ann E. McCullough, Bhavika K. Patel, Jeffrey N. Weitzel, Thomas P. Slavin, Carlos Caldas, Barbara A. Pockaj, and Muhammed Murtaza. Personalized Circulating Tumor DNA Analysis to Detect Residual Disease After NeoAdjuvant Therapy in Breast Cancer. Sci Transl Med. 2019 August 07; 11(504) (doi:10.1126/scitranslmed.aax7392).
6. Yves Konigshofer, Mathew G. Butler, Jessica Dickens, Omoshile Clement, Andrew Anfora, Dan Brudewsky, Bharathi Anekela, and Russell Garlick, Reference Materials for Measurable Residual Disease (MRD) Monitoring, Poster# 1978, 2020 AACR Virtual Meeting II, June 2020.
7. Mathew G. Butler, Yves Konigshofer, Omoshile Clement, Andrew Anfora, Dan Brudzewsky, Bharathi Anekella, and Russell Garlick, Development of Reference Materials for Blood Tumor Mutational Burden (bTMB), Poster# 1982, 2020 AACR Virtual Meeting II, June 2020.
8. Eun-Ang Rabier-Moreau; Guillem Portella; Matthew Butler; Yves Konigshofer; James Hadfield; Development and validation of blood tumor mutational burden reference standards, 2020 SITC Conference, Abstract #60, Nov 2020.
9. Yves Konigshofer, Matthew G. Butler, Jessica L. Dickens, Omoshile Clement, Bharathi Anekella, and Russell K. Garlick, Reference Materials for Measurable Residual Disease (MRD) Monitoring in Circulating Cell-Free DNA (ccfDNA), Poster TT12: 2020 AMP Meeting, Nov 2020.


